Abstract
The DNA Methyltransferase 3A (DNMT3A) gene is recurrently mutated in a large spectrum of hematologic malignancies, including acute myeloid leukemia (AML). About 25% of adult AML patients carry mutations in DNMT3A and these mutations are generally associated with poor prognosis. DNMT3A mutations have been also associated with aged-related clonal hematopoiesis of indeterminate potential (CHIP). The high prevalence of DNMT3A somatic mutations in AML and CHIP implies that cells with mutated DNMT3A have a competitive advantage over wild-type (WT) cells, resulting in clonal expansion. However, the downstream molecular mechanisms that underlie this phenotype are not clear.
Tatton-Brown-Raman syndrome (TBRS) is a rare genetic disorder caused by heterozygous germline mutations in DNMT3A, characterized by overgrowth and developmental delay. In one particular family, a group of 4 children out of 12 were diagnosed with TBRS and were found to be heterozygous carriers of DNMT3A-R771Q mutation (DNMT3AR771Q) inherited from their mosaic father. Thus, this individual provides a unique opportunity to study the long-term consequences of DNMT3A mutations, as he harbors both WT and mutant cells.
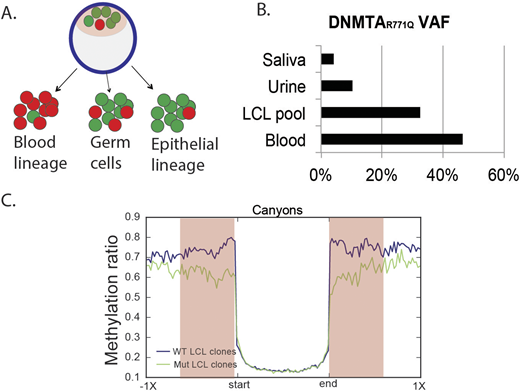
From this mosaic individual, we generated lymphoblastoid cell lines (LCLs) from the peripheral blood (PB) and measured DNMT3AR771Q variant allele frequency (VAF) in the LCL pool as well as in PB, saliva and urine, all collected at the same time. Strikingly, DNMT3AR771Q VAF in the LCL pool and in PB was substantially higher than in saliva and urine (respectively 30%, 45%, 10%, 4%), implying that levels of DNMT3A mosaicism are tissue-specific and that cells with mutated DNMT3A tend to expand in the blood but not in epithelia (figure 1A and figure1B).
One hypothesis for the prevalence of DNMT3A mutations in AML is that its loss reduces the effectiveness of DNA repair leading to increased mutational rates. In order to test this, we compared the mutational loads in individual LCL clones that were WT or DNMT3A mutant using whole genome sequencing. Surprisingly, no clear differences were observed between WT and DNMT3AR771Q mutant cells, indicating that clonal expansion is unlikely to be secondary to a general increase in mutational burden.
To explore the impact of DNMT3AR771Q mutation on DNA methylation, we performed whole-genome bisulfite sequencing (WGBS) on two WT and two DNMT3AR771Q LCL clones. We identified ~31,500 differentially methylated regions (DMRs) between WT and mutant clones, with the majority of DMRs being hypomethylated. Hypomethylated DMRs were associated with gene regulatory regions, mainly promoters and enhancer regions. These data suggest that the DNMT3AR771Q mutation affects DNA methylation setting at genomic regions that can directly affect transcription.
Canyons are large genomic regions of low methylation that often occur around master regulators such as homeobox-containing genes. We previously showed in mice that DNMT3A regulates DNA methylation at canyon edges, with loss of DNMT3A resulting in canyon expansion. In agreement, DNMT3AR771Q mutant clones displayed larger canyons, particularly at loci marked by H3K27Ac and H3K4me3 (figure 1C). Gene Ontology analysis of genes falling into expanded canyons showed a significant enrichment for leukemia and stem cell-related genes, including members of the HOX family. RNAseq analysis of DNMT3AR771Q mutant LCL clones confirmed the upregulation of key cancer-associated genes. These data suggest that DNMT3A mutations may promote clonal expansion through hypomethylation and overexpression of stem cell and cancer-related genes
In conclusion, by comparing WT and DNMT3Amutant LCL clones generated from the same individual, we show that DNMT3A mutations lead to significant hypomethylation and overexpression of key cancer-associated genes. Further studies on specific target genes will reveal critical pathways responsible for the clonal expansion of cells with mutated DNMT3A, paving the way for the development of new therapeutic strategies for malignancies with mutated DNMT3A.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.